I was reading the Evo paper, and it referred the Evo model as a foundation model (基础模型). I had to look up what that means.
In the context of this paper, a foundation model refers to a large, general-purpose machine learning model that is trained on vast amounts of data and can be adapted (or fine-tuned) for a wide range of downstream tasks. Foundation models are designed to capture broad patterns and relationships in the data, making them highly versatile and powerful tools for various applications.
Key Characteristics of Foundation Models:
- Large-Scale Training: Foundation models are trained on massive datasets, often using unsupervised or self-supervised learning techniques.
- General-Purpose: They are not task-specific but are designed to learn general representations of the data (e.g., language, images, or biological sequences).
- Transfer Learning: Once trained, foundation models can be fine-tuned or adapted to specific tasks with relatively little additional data.
- Versatility: They can be applied across multiple domains and tasks, often outperforming specialized models.
Examples of Foundation Models:
- Natural Language Processing (NLP):
- GPT (Generative Pre-trained Transformer):
- Developed by OpenAI, GPT models (e.g., GPT-3, GPT-4) are trained on vast amounts of text data and can perform tasks like text generation, translation, summarization, and question answering.
- BERT (Bidirectional Encoder Representations from Transformers):
- Developed by Google, BERT is trained to understand the context of words in a sentence and is used for tasks like sentiment analysis, named entity recognition, and question answering.
- T5 (Text-To-Text Transfer Transformer):
- Developed by Google, T5 treats all NLP tasks as a text-to-text problem, making it highly flexible for tasks like translation, summarization, and classification.
- GPT (Generative Pre-trained Transformer):
- Computer Vision:
- CLIP (Contrastive Language–Image Pretraining):
- Developed by OpenAI, CLIP connects images and text, enabling tasks like zero-shot image classification and image-text retrieval.
- DALL·E:
- Also developed by OpenAI, DALL·E generates images from textual descriptions, demonstrating the ability to combine vision and language understanding.
- CLIP (Contrastive Language–Image Pretraining):
- Biology and Bioinformatics:
- Protein Models:
- AlphaFold: Developed by DeepMind, AlphaFold predicts protein structures from amino acid sequences, revolutionizing structural biology.
- ESM (Evolutionary Scale Modeling): Developed by Meta AI, ESM models are trained on protein sequences to predict structure, function, and evolutionary relationships.
- DNA Models:
- DNABERT
- NT (Nucleotide Transfoermer)
- Evo: Evo is a foundation model designed to capture the multimodality of the central dogma (DNA → RNA → protein) and the multiscale nature of evolution. It can likely be applied to tasks like gene function prediction, protein design, and evolutionary analysis. Evo2 is just released and eukaryotic genomes are included in the training this time.
- Protein Models:
- Multimodal Models:
- Flamingo:
- Developed by DeepMind, Flamingo combines vision and language understanding, enabling tasks like image captioning and visual question answering.
- Gato:
- Also developed by DeepMind, Gato is a general-purpose model capable of performing tasks across multiple domains, including text, images, and robotics.
- Flamingo:
Why Foundation Models Are Important:
- Efficiency: Instead of training a new model from scratch for every task, foundation models can be fine-tuned with minimal additional data and computation.
- Performance: Foundation models often achieve state-of-the-art performance on a wide range of tasks due to their large-scale training and generalization capabilities.
- Innovation: They enable new applications and discoveries by providing a powerful base for further research and development.
Evo as a Foundation Model:
In the case of Evo, it is designed to capture two key aspects of biology:
- Multimodality of the Central Dogma: Evo can handle the flow of genetic information from DNA to RNA to proteins, integrating multiple biological modalities.
- Multiscale Nature of Evolution: Evo can analyze evolutionary patterns at different scales, from molecular scales to systems scales(interaction between different modality of molecules) and entire genomes (see their figure from the paper below).
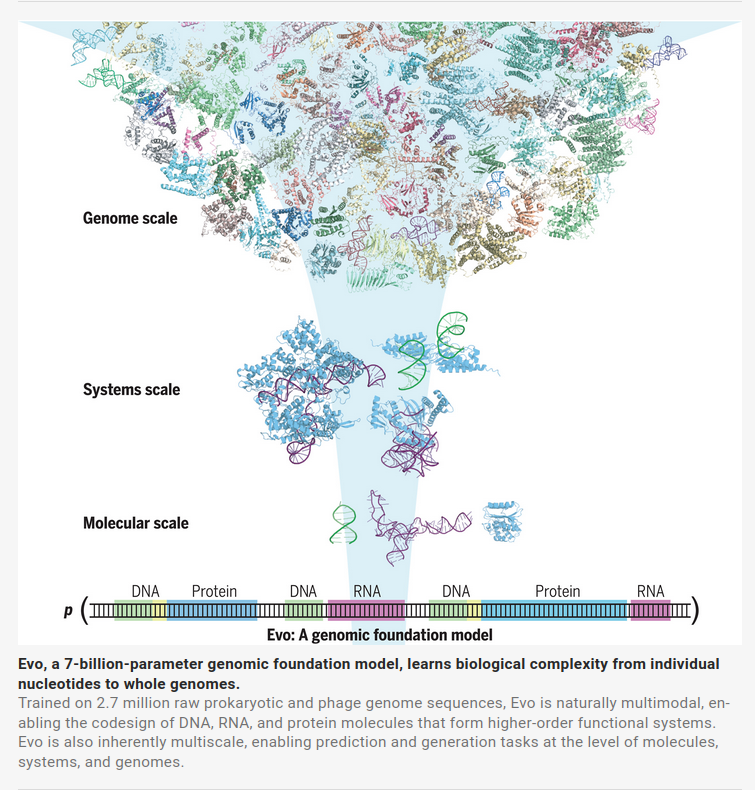
As a foundation model, Evo can be fine-tuned for various biological tasks, such as predicting gene functions, designing proteins, or analyzing evolutionary relationships, making it a versatile tool for computational biology.
Update from Evo2
-
Evo is trained on prokaryotes and phage genomes. Evo2 is trained on “a highly curated genomic atlas spanning all domains of life”.
-
Pretraining data set: 300 billion nt (from 2.7 million genomes) vs. 9.3 (abstract) or 8.84 (methods) trillion nt. openGenome2 (the one Evo 2 was trained on) included a 33% expansion of representative prokaryotic genomes from 85,205 to 113,379 (357 billion nucleotides), a total of 6.98 trillion nucleotides from eukaryotic genomes, 854 billion nucleotides of non-redundant metagenomic sequencing data, 2.82 billion nucleotides of organelle genomes, and 602 billion nucleotides of subsets of eukaryotic sequence data to focus on likely functional regions of the genomes by focusing on different windows around coding genes. This means eukaryotic genomes takes about 80% of the pretraining dataset.
Let’s take a closer look on how the eukaryotic genomes were chosen:
vs. 9.3 trillion DNA base pairs.
- Model parameter size: 7B(Evo) vs 7B and 40B (Evo2). The differences between the two Evo2 model is that the 7B parameters trained on 2.4 trillion tokens and a full version at 40B parameters trained on 9.3 trillion tokens. Note that GPT3 has 175B.
- Token context window1 million: 131Kb vs 1Mb, both at single-nucleotide resolution. This number is 2048 for GPT3.
- Evo 2 learns from DNA sequence alone to accurately predict the functional impacts of genetic variation.